Single stack tutorial based on the 3D Burgers + tracer equations
This tutorial implements the Burgers equations with a tracer field in a single element stack. The flow is initialized with a horizontally uniform profile of horizontal velocity and uniform initial temperature. The fluid is heated from the bottom surface. Gaussian noise is imposed to the horizontal velocity field at each node at the start of the simulation. The tutorial demonstrates how to
- Initialize a
BalanceLawin a single stack configuration; - Return the horizontal velocity field to a given profile (e.g., large-scale advection);
- Remove any horizontal inhomogeneities or noise from the flow.
The second and third bullet points are demonstrated imposing Rayleigh friction, horizontal diffusion and 2D divergence damping to the horizontal momentum prognostic equation.
Equations solved in balance law form:
\[\begin{align} \frac{∂ ρ}{∂ t} =& - ∇ ⋅ (ρ\mathbf{u}) \\ \frac{∂ ρ\mathbf{u}}{∂ t} =& - ∇ ⋅ (-μ ∇\mathbf{u}) - ∇ ⋅ (ρ\mathbf{u} \mathbf{u}') - γ[ (ρ\mathbf{u}-ρ̄\mathbf{ū}) - (ρ\mathbf{u}-ρ̄\mathbf{ū})⋅ẑ ẑ] - ν_d ∇_h (∇_h ⋅ ρ\mathbf{u}) \\ \frac{∂ ρcT}{∂ t} =& - ∇ ⋅ (-α ∇ρcT) - ∇ ⋅ (\mathbf{u} ρcT) \end{align}\]
Boundary conditions:
\[\begin{align} z_{\mathrm{min}}: & ρ = 1 \\ z_{\mathrm{min}}: & ρ\mathbf{u} = \mathbf{0} \\ z_{\mathrm{min}}: & ρcT = ρc T_{\mathrm{fixed}} \\ z_{\mathrm{max}}: & ρ = 1 \\ z_{\mathrm{max}}: & ρ\mathbf{u} = \mathbf{0} \\ z_{\mathrm{max}}: & -α∇ρcT = 0 \end{align}\]
where
- $t$ is time
- $ρ$ is the density
- $\mathbf{u}$ is the velocity (vector)
- $\mathbf{ū}$ is the horizontally averaged velocity (vector)
- $μ$ is the dynamic viscosity tensor
- $γ$ is the Rayleigh friction frequency
- $ν_d$ is the horizontal divergence damping coefficient
- $T$ is the temperature
- $α$ is the thermal diffusivity tensor
- $c$ is the heat capacity
- $ρcT$ is the thermal energy
Solving these equations is broken down into the following steps:
- Preliminary configuration
- PDEs
- Space discretization
- Time discretization
- Solver hooks / callbacks
- Solve
- Post-processing
Preliminary configuration
Loading code
First, we'll load our pre-requisites
- load external packages:
using MPI
using Distributions
using OrderedCollections
using Plots
using StaticArrays
using LinearAlgebra: Diagonal, tr- load CLIMAParameters and set up to use it:
using CLIMAParameters
struct EarthParameterSet <: AbstractEarthParameterSet end
const param_set = EarthParameterSet()Main.##346.EarthParameterSet()- load necessary ClimateMachine modules:
using ClimateMachine
using ClimateMachine.Mesh.Topologies
using ClimateMachine.Mesh.Grids
using ClimateMachine.Writers
using ClimateMachine.DGMethods
using ClimateMachine.DGMethods.NumericalFluxes
using ClimateMachine.BalanceLaws:
BalanceLaw, Prognostic, Auxiliary, Gradient, GradientFlux, parameter_set
using ClimateMachine.Mesh.Geometry: LocalGeometry
using ClimateMachine.MPIStateArrays
using ClimateMachine.GenericCallbacks
using ClimateMachine.ODESolvers
using ClimateMachine.VariableTemplates
using ClimateMachine.SingleStackUtils- import necessary ClimateMachine modules: (
importing enables us to
provide implementations of these structs/methods)
using ClimateMachine.Orientations:
Orientation,
FlatOrientation,
init_aux!,
vertical_unit_vector,
projection_tangential
import ClimateMachine.BalanceLaws:
vars_state,
source!,
flux_second_order!,
flux_first_order!,
compute_gradient_argument!,
compute_gradient_flux!,
init_state_auxiliary!,
init_state_prognostic!,
BoundaryCondition,
boundary_conditions,
boundary_state!Initialization
Define the float type (Float64 or Float32)
const FT = Float64;Initialize ClimateMachine for CPU.
ClimateMachine.init(; disable_gpu = true);
const clima_dir = dirname(dirname(pathof(ClimateMachine)));[1638959905.719476] [hpc-92-37:26678:0] ib_verbs.h:84 UCX ERROR ibv_exp_query_device(mlx5_0) returned 38: No space left on device
Load some helper functions for plotting
include(joinpath(clima_dir, "docs", "plothelpers.jl"));Define the set of Partial Differential Equations (PDEs)
Define the model
Model parameters can be stored in the particular BalanceLaw, in this case, the BurgersEquation:
Base.@kwdef struct BurgersEquation{FT, APS, O} <: BalanceLaw
"Parameters"
param_set::APS
"Orientation model"
orientation::O
"Heat capacity"
c::FT = 1
"Vertical dynamic viscosity"
μv::FT = 1e-4
"Horizontal dynamic viscosity"
μh::FT = 1
"Vertical thermal diffusivity"
αv::FT = 1e-2
"Horizontal thermal diffusivity"
αh::FT = 1
"IC Gaussian noise standard deviation"
σ::FT = 5e-2
"Rayleigh damping"
γ::FT = 5
"Domain height"
zmax::FT = 1
"Initial conditions for temperature"
initialT::FT = 295.15
"Bottom boundary value for temperature (Dirichlet boundary conditions)"
T_bottom::FT = 300.0
"Top flux (α∇ρcT) at top boundary (Neumann boundary conditions)"
flux_top::FT = 0.0
"Divergence damping coefficient (horizontal)"
νd::FT = 1
endCreate an instance of the BurgersEquation:
orientation = FlatOrientation()
m = BurgersEquation{FT, typeof(param_set), typeof(orientation)}(
param_set = param_set,
orientation = orientation,
);This model dictates the flow control, using Dynamic Multiple Dispatch, for which kernels are executed.
Define the variables
All of the methods defined in this section were imported in Loading code to let us provide implementations for our BurgersEquation as they will be used by the solver.
Specify auxiliary variables for BurgersEquation
function vars_state(m::BurgersEquation, st::Auxiliary, FT)
@vars begin
coord::SVector{3, FT}
orientation::vars_state(m.orientation, st, FT)
end
endvars_state (generic function with 152 methods)Specify prognostic variables, the variables solved for in the PDEs, for BurgersEquation
vars_state(::BurgersEquation, ::Prognostic, FT) =
@vars(ρ::FT, ρu::SVector{3, FT}, ρcT::FT);Specify state variables whose gradients are needed for BurgersEquation
vars_state(::BurgersEquation, ::Gradient, FT) =
@vars(u::SVector{3, FT}, ρcT::FT, ρu::SVector{3, FT});Specify gradient variables for BurgersEquation
vars_state(::BurgersEquation, ::GradientFlux, FT) = @vars(
μ∇u::SMatrix{3, 3, FT, 9},
α∇ρcT::SVector{3, FT},
νd∇D::SMatrix{3, 3, FT, 9}
);Define the compute kernels
Specify the initial values in aux::Vars, which are available in init_state_prognostic!. Note that
- this method is only called at
t=0. aux.coordis available here because we've specifiedcoordinvars_state(m, aux, FT).
function nodal_init_state_auxiliary!(
m::BurgersEquation,
aux::Vars,
tmp::Vars,
geom::LocalGeometry,
)
aux.coord = geom.coord
end;init_aux! initializes the auxiliary gravitational potential field needed for vertical projections
function init_state_auxiliary!(
m::BurgersEquation,
state_auxiliary::MPIStateArray,
grid,
direction,
)
init_aux!(m, m.orientation, state_auxiliary, grid, direction)
init_state_auxiliary!(
m,
nodal_init_state_auxiliary!,
state_auxiliary,
grid,
direction,
)
end;Specify the initial values in state::Vars. Note that
- this method is only called at
t=0. state.ρ,state.ρuandstate.ρcTare available here because we've specifiedρ,ρuandρcTinvars_state(m, state, FT).
function init_state_prognostic!(
m::BurgersEquation,
state::Vars,
aux::Vars,
localgeo,
t::Real,
)
z = aux.coord[3]
ε1 = rand(Normal(0, m.σ))
ε2 = rand(Normal(0, m.σ))
state.ρ = 1
ρu = 1 - 4 * (z - m.zmax / 2)^2 + ε1
ρv = 1 - 4 * (z - m.zmax / 2)^2 + ε2
ρw = 0
state.ρu = SVector(ρu, ρv, ρw)
state.ρcT = state.ρ * m.c * m.initialT
end;The remaining methods, defined in this section, are called at every time-step in the solver by the BalanceLaw framework.
Since we have second-order fluxes, we must tell ClimateMachine to compute the gradient of ρcT, u and ρu. Here, we specify how ρcT, u and ρu are computed. Note that e.g. transform.ρcT is available here because we've specified ρcT in vars_state(m, ::Gradient, FT).
function compute_gradient_argument!(
m::BurgersEquation,
transform::Vars,
state::Vars,
aux::Vars,
t::Real,
)
transform.ρcT = state.ρcT
transform.u = state.ρu / state.ρ
transform.ρu = state.ρu
end;Specify where in diffusive::Vars to store the computed gradient from compute_gradient_argument!. Note that:
diffusive.μ∇uis available here because we've specifiedμ∇uinvars_state(m, ::GradientFlux, FT).∇transform.uis available here because we've specifieduinvars_state(m, ::Gradient, FT).diffusive.μ∇uis built using an anisotropic diffusivity tensor.- The
divergencemay be computed from the trace of tensor∇ρu.
function compute_gradient_flux!(
m::BurgersEquation{FT},
diffusive::Vars,
∇transform::Grad,
state::Vars,
aux::Vars,
t::Real,
) where {FT}
param_set = parameter_set(m)
∇ρu = ∇transform.ρu
ẑ = vertical_unit_vector(m.orientation, param_set, aux)
divergence = tr(∇ρu) - ẑ' * ∇ρu * ẑ
diffusive.α∇ρcT = Diagonal(SVector(m.αh, m.αh, m.αv)) * ∇transform.ρcT
diffusive.μ∇u = Diagonal(SVector(m.μh, m.μh, m.μv)) * ∇transform.u
diffusive.νd∇D =
Diagonal(SVector(m.νd, m.νd, FT(0))) *
Diagonal(SVector(divergence, divergence, FT(0)))
end;Introduce Rayleigh friction towards a target profile as a source. Note that:
- Rayleigh damping is only applied in the horizontal using the
projection_tangentialmethod.
function source!(
m::BurgersEquation{FT},
source::Vars,
state::Vars,
diffusive::Vars,
aux::Vars,
args...,
) where {FT}
param_set = parameter_set(m)
ẑ = vertical_unit_vector(m.orientation, param_set, aux)
z = aux.coord[3]
ρ̄ū =
state.ρ * SVector{3, FT}(
0.5 - 2 * (z - m.zmax / 2)^2,
0.5 - 2 * (z - m.zmax / 2)^2,
0.0,
)
ρu_p = state.ρu - ρ̄ū
source.ρu -=
m.γ * projection_tangential(m.orientation, param_set, aux, ρu_p)
end;Compute advective flux. Note that:
state.ρuis available here because we've specifiedρuinvars_state(m, state, FT).
function flux_first_order!(
m::BurgersEquation,
flux::Grad,
state::Vars,
aux::Vars,
t::Real,
_...,
)
flux.ρ = state.ρu
u = state.ρu / state.ρ
flux.ρu = state.ρu * u'
flux.ρcT = u * state.ρcT
end;Compute diffusive flux (e.g. $F(μ, \mathbf{u}, t) = -μ∇\mathbf{u}$ in the original PDE). Note that:
diffusive.μ∇uis available here because we've specifiedμ∇uinvars_state(m, ::GradientFlux, FT).- The divergence gradient can be written as a diffusive flux using a divergence diagonal tensor.
function flux_second_order!(
m::BurgersEquation,
flux::Grad,
state::Vars,
diffusive::Vars,
hyperdiffusive::Vars,
aux::Vars,
t::Real,
)
flux.ρcT -= diffusive.α∇ρcT
flux.ρu -= diffusive.μ∇u
flux.ρu -= diffusive.νd∇D
end;Boundary conditions
Second-order terms in our equations, $∇⋅(G)$ where $G = μ∇\mathbf{u}$, are internally reformulated to first-order unknowns. Boundary conditions must be specified for all unknowns, both first-order and second-order unknowns which have been reformulated.
struct TopBC <: BoundaryCondition end;
struct BottomBC <: BoundaryCondition end;
boundary_conditions(::BurgersEquation) = (BottomBC(), TopBC());The boundary conditions for ρ, ρu and ρcT (first order unknowns)
function boundary_state!(
nf,
bc::BottomBC,
m::BurgersEquation,
state⁺::Vars,
aux⁺::Vars,
n⁻,
_...,
)
state⁺.ρ = 1
state⁺.ρu = SVector(0, 0, 0)
state⁺.ρcT = state⁺.ρ * m.c * m.T_bottom
end;
function boundary_state!(
nf,
bc::TopBC,
m::BurgersEquation,
state⁺::Vars,
aux⁺::Vars,
n⁻,
_...,
)
state⁺.ρ = 1
state⁺.ρu = SVector(0, 0, 0)
end;The boundary conditions for ρ, ρu and ρcT are specified here for second-order unknowns
function boundary_state!(
nf,
bc::BottomBC,
m::BurgersEquation,
state⁺::Vars,
diff⁺::Vars,
hyperdiff⁺::Vars,
aux⁺::Vars,
n⁻,
_...,
)
state⁺.ρ = 1
state⁺.ρu = SVector(0, 0, 0)
state⁺.ρcT = state⁺.ρ * m.c * m.T_bottom
end;
function boundary_state!(
nf,
bc::TopBC,
m::BurgersEquation,
state⁺::Vars,
diff⁺::Vars,
hyperdiff⁺::Vars,
aux⁺::Vars,
n⁻,
_...,
)
state⁺.ρ = 1
state⁺.ρu = SVector(0, 0, 0)
diff⁺.α∇ρcT = -n⁻ * m.flux_top
end;Spatial discretization
Prescribe polynomial order of basis functions in finite elements
N_poly = 5;Specify the number of vertical elements
nelem_vert = 10;Specify the domain height
zmax = m.zmax;Establish a ClimateMachine single stack configuration
driver_config = ClimateMachine.SingleStackConfiguration(
"BurgersEquation",
N_poly,
nelem_vert,
zmax,
param_set,
m,
numerical_flux_first_order = CentralNumericalFluxFirstOrder(),
);ClimateMachine.array_type() = Array
┌ Info: Model composition
│ param_set = Main.##346.EarthParameterSet()
│ orientation = ClimateMachine.Orientations.FlatOrientation()
│ c = 1.0
│ μv = 0.0001
│ μh = 1.0
│ αv = 0.01
│ αh = 1.0
│ σ = 0.05
│ γ = 5.0
│ zmax = 1.0
│ initialT = 295.15
│ T_bottom = 300.0
│ flux_top = 0.0
│ νd = 1.0
└ @ ClimateMachine /central/scratch/climaci/climatemachine-docs/1430/climatemachine-docs/src/Driver/driver_configs.jl:188
┌ Info: Defining `prognostic_vars` and
│ `eq_tends` for BurgersEquation will
│ enable printing a table of tendencies.
└ @ ClimateMachine.BalanceLaws /central/scratch/climaci/climatemachine-docs/1430/climatemachine-docs/src/BalanceLaws/show_tendencies.jl:65
┌ Info: Establishing single stack configuration for BurgersEquation
│ precision = Float64
│ horiz polynomial order = 5
│ vert polynomial order = 5
│ domain_min = 0.00 m, 0.00 m, 0.00 m
│ domain_max = 1.00 m, 1.00 m, 1.00 m
│ # vert elems = 10
│ MPI ranks = 1
│ min(Δ_horz) = 0.12 m
│ min(Δ_vert) = 0.01 m
└ @ ClimateMachine /central/scratch/climaci/climatemachine-docs/1430/climatemachine-docs/src/Driver/driver_configs.jl:612
Time discretization
Specify simulation time (SI units)
t0 = FT(0);
timeend = FT(1);We'll define the time-step based on the Fourier number and the Courant number of the flow
Δ = min_node_distance(driver_config.grid)
given_Fourier = FT(0.5);
Fourier_bound = given_Fourier * Δ^2 / max(m.αh, m.μh, m.νd);
Courant_bound = FT(0.5) * Δ;
dt = min(Fourier_bound, Courant_bound)6.899875101735774e-5Configure a ClimateMachine solver.
This initializes the state vector and allocates memory for the solution in space (dg has the model m, which describes the PDEs as well as the function used for initialization). This additionally initializes the ODE solver, by default an explicit Low-Storage Runge-Kutta method.
solver_config =
ClimateMachine.SolverConfiguration(t0, timeend, driver_config, ode_dt = dt);┌ Info: Initializing BurgersEquation
└ @ ClimateMachine /central/scratch/climaci/climatemachine-docs/1430/climatemachine-docs/src/Driver/solver_configs.jl:185
Inspect the initial conditions for a single nodal stack
Let's export plots of the initial state
output_dir = @__DIR__;
mkpath(output_dir);
z_scale = 100 # convert from meters to cm
z_key = "z"
z_label = "z [cm]"
z = get_z(driver_config.grid; z_scale = z_scale)
state_vars = get_vars_from_nodal_stack(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
);Create an array to store the solution:
state_data = Dict[state_vars] # store initial condition at ``t=0``
time_data = FT[0] # store time data1-element Array{Float64,1}:
0.0Generate plots of initial conditions for the southwest nodal stack
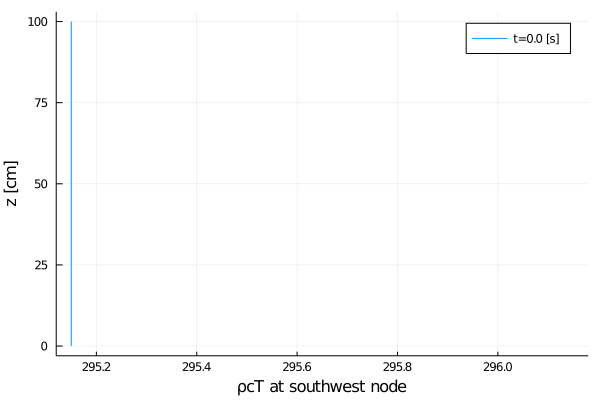
export_plot(
z,
time_data,
state_data,
("ρcT",),
joinpath(output_dir, "initial_condition_T_nodal.png");
xlabel = "ρcT at southwest node",
ylabel = z_label,
);
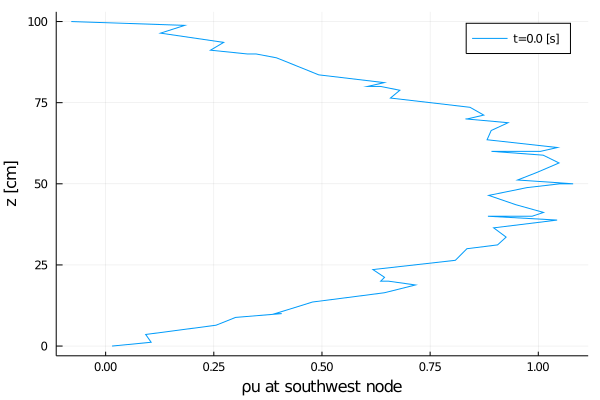
export_plot(
z,
time_data,
state_data,
("ρu[1]",),
joinpath(output_dir, "initial_condition_u_nodal.png");
xlabel = "ρu at southwest node",
ylabel = z_label,
);
export_plot(
z,
time_data,
state_data,
("ρu[2]",),
joinpath(output_dir, "initial_condition_v_nodal.png");
xlabel = "ρv at southwest node",
ylabel = z_label,
);
Inspect the initial conditions for the horizontal averages
Horizontal statistics of variables
state_vars_var = get_horizontal_variance(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
);
state_vars_avg = get_horizontal_mean(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
);
data_avg = Dict[state_vars_avg]
data_var = Dict[state_vars_var]
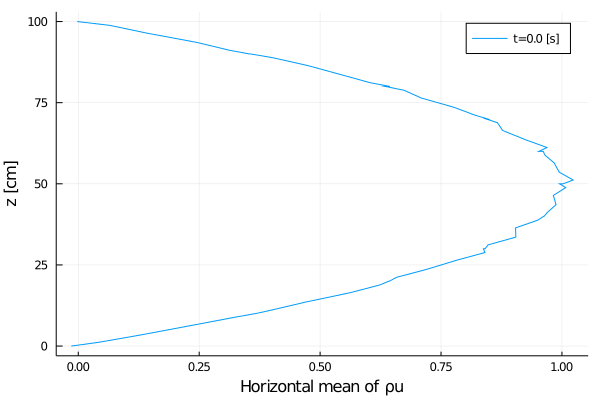
export_plot(
z,
time_data,
data_avg,
("ρu[1]",),
joinpath(output_dir, "initial_condition_avg_u.png");
xlabel = "Horizontal mean of ρu",
ylabel = z_label,
);
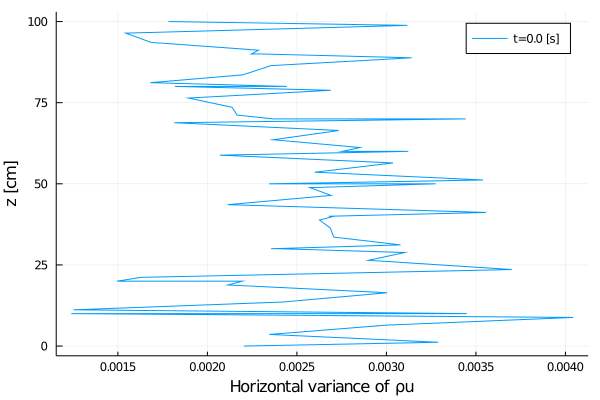
export_plot(
z,
time_data,
data_var,
("ρu[1]",),
joinpath(output_dir, "initial_condition_variance_u.png");
xlabel = "Horizontal variance of ρu",
ylabel = z_label,
);
Solver hooks / callbacks
Define the number of outputs from t0 to timeend
const n_outputs = 5;
const every_x_simulation_time = timeend / n_outputs;Create a dictionary for z coordinate (and convert to cm) NCDatasets IO:
dims = OrderedDict(z_key => collect(z));Create dictionaries to store outputs:
data_var = Dict[Dict([k => Dict() for k in 0:n_outputs]...),]
data_var[1] = state_vars_var
data_avg = Dict[Dict([k => Dict() for k in 0:n_outputs]...),]
data_avg[1] = state_vars_avg
data_nodal = Dict[Dict([k => Dict() for k in 0:n_outputs]...),]
data_nodal[1] = state_varsOrderedCollections.OrderedDict{Any,Any} with 5 entries:
"ρ" => [1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0]
"ρu[1]" => [0.0148644, 0.105171, 0.0925204, 0.255171, 0.300145, 0.407007, 0.391372, 0.420351, 0.478166, 0.644204, 0.715958, 0.653591, 0.635729, 0.644587, 0.617726, 0.80829, 0.826228, 0.83503, 0.83508, 0.905845, 0.92563, 0.896795, 1.04351, 0.883575, 0.985542, 1.01201, 0.947663, 0.885378, 0.973802, 1.05005, 1.08053, 0.952127, 0.998048, 1.04788, 1.01118, 0.891746, 1.00518, 1.04406, 0.88164, 0.891114, 0.930119, 0.831635, 0.835757, 0.87394, 0.842159, 0.658266, 0.679976, 0.63545, 0.605204, 0.643828, 0.492485, 0.439529, 0.394777, 0.348156, 0.328265, 0.242296, 0.272756, 0.126778, 0.18266, -0.0794399]
"ρu[2]" => [0.0191198, -0.0199116, 0.0893337, 0.273913, 0.334051, 0.356575, 0.329935, 0.431803, 0.469631, 0.530675, 0.574463, 0.672502, 0.644464, 0.662988, 0.798274, 0.736105, 0.863029, 0.840923, 0.948651, 0.759241, 0.864755, 0.921254, 1.05263, 0.877743, 0.943961, 0.843966, 0.929062, 0.972561, 0.970572, 0.964499, 0.950011, 1.02794, 0.94174, 1.00048, 1.13197, 0.933958, 0.914696, 0.946138, 0.845406, 0.792641, 0.874587, 0.819106, 0.900191, 0.878725, 0.793918, 0.676004, 0.682186, 0.651182, 0.59164, 0.524234, 0.61556, 0.352861, 0.442435, 0.426561, 0.357215, 0.410848, 0.262358, 0.137709, 0.166466, -0.0862527]
"ρu[3]" => [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]
"ρcT" => [295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15, 295.15]The ClimateMachine's time-steppers provide hooks, or callbacks, which allow users to inject code to be executed at specified intervals. In this callback, the state variables are collected, combined into a single OrderedDict and written to a NetCDF file (for each output step).
callback = GenericCallbacks.EveryXSimulationTime(every_x_simulation_time) do
state_vars_var = get_horizontal_variance(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
)
state_vars_avg = get_horizontal_mean(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
)
state_vars = get_vars_from_nodal_stack(
driver_config.grid,
solver_config.Q,
vars_state(m, Prognostic(), FT),
i = 1,
j = 1,
)
push!(data_var, state_vars_var)
push!(data_avg, state_vars_avg)
push!(data_nodal, state_vars)
push!(time_data, gettime(solver_config.solver))
nothing
end;Solve
This is the main ClimateMachine solver invocation. While users do not have access to the time-stepping loop, code may be injected via user_callbacks, which is a Tuple of GenericCallbacks.
ClimateMachine.invoke!(solver_config; user_callbacks = (callback,))1.0018567587564864Post-processing
Our solution has now been calculated and exported to NetCDF files in output_dir.
Let's plot the horizontal statistics of ρu and ρcT, as well as the evolution of ρu for the southwest nodal stack:
export_plot(
z,
time_data,
data_avg,
("ρu[1]"),
joinpath(output_dir, "solution_vs_time_u_avg.png");
xlabel = "Horizontal mean of ρu",
ylabel = z_label,
);
export_plot(
z,
time_data,
data_var,
("ρu[1]"),
joinpath(output_dir, "variance_vs_time_u.png");
xlabel = "Horizontal variance of ρu",
ylabel = z_label,
);
export_plot(
z,
time_data,
data_avg,
("ρcT"),
joinpath(output_dir, "solution_vs_time_T_avg.png");
xlabel = "Horizontal mean of ρcT",
ylabel = z_label,
);
export_plot(
z,
time_data,
data_var,
("ρcT"),
joinpath(output_dir, "variance_vs_time_T.png");
xlabel = "Horizontal variance of ρcT",
ylabel = z_label,
);
export_plot(
z,
time_data,
data_nodal,
("ρu[1]"),
joinpath(output_dir, "solution_vs_time_u_nodal.png");
xlabel = "ρu at southwest node",
ylabel = z_label,
);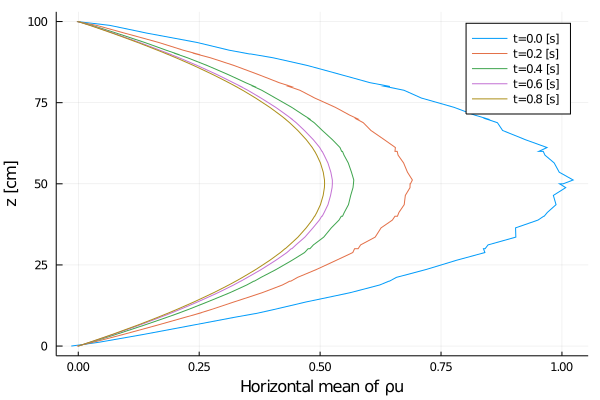
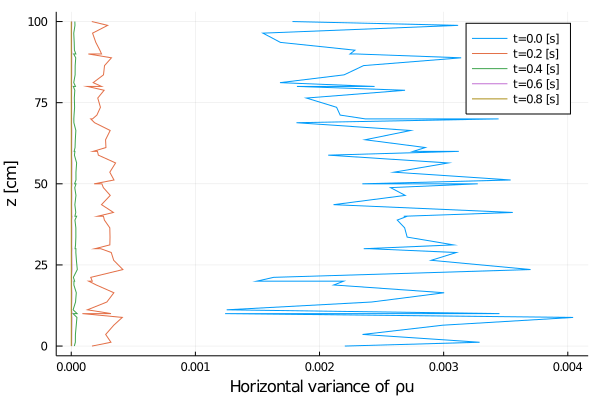
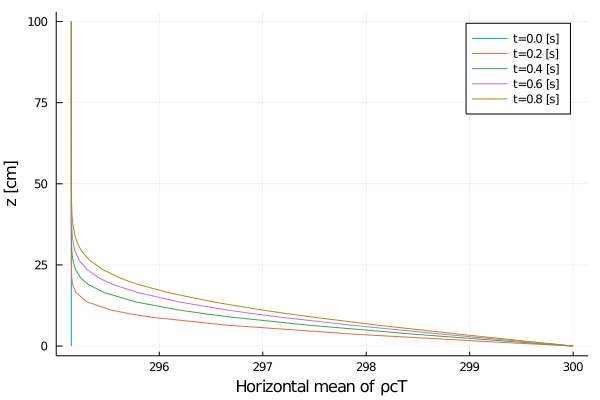
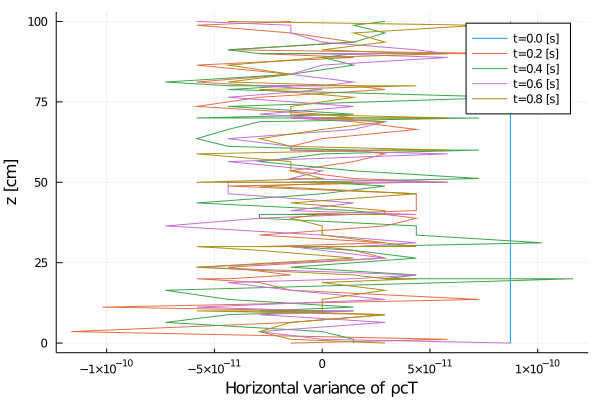
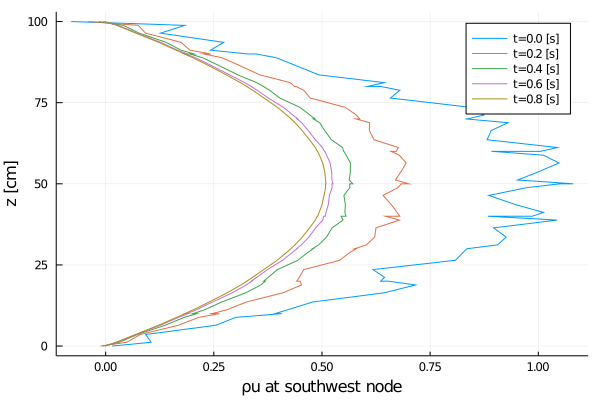
Rayleigh friction returns the horizontal velocity to the objective profile on the timescale of the simulation (1 second), since γ∼1. The horizontal viscosity and 2D divergence damping act to reduce the horizontal variance over the same timescale. The initial Gaussian noise is propagated to the temperature field through advection. The horizontal diffusivity acts to reduce this ρcT variance in time, although in a longer timescale.
To run this file, and inspect the solution, include this tutorial in the Julia REPL with:
include(joinpath("tutorials", "Atmos", "burgers_single_stack.jl"))This page was generated using Literate.jl.