Baroclinic adjustment
In this example, we simulate the evolution and equilibration of a baroclinically unstable front.
Install dependencies
First let's make sure we have all required packages installed.
using Pkg
pkg"add Oceananigans, CairoMakie"using Oceananigans
using Oceananigans.Units
using Random
Random.seed!(8675309) # for reproducible resultsRandom.TaskLocalRNG()Grid
We use a three-dimensional channel that is periodic in the x direction:
Lx = 1000kilometers # east-west extent [m]
Ly = 1000kilometers # north-south extent [m]
Lz = 1kilometers # depth [m]
grid = RectilinearGrid(size = (48, 48, 8),
x = (0, Lx),
y = (-Ly/2, Ly/2),
z = (-Lz, 0),
topology = (Periodic, Bounded, Bounded))48×48×8 RectilinearGrid{Float64, Periodic, Bounded, Bounded} on CPU with 3×3×3 halo
├── Periodic x ∈ [0.0, 1.0e6) regularly spaced with Δx=20833.3
├── Bounded y ∈ [-500000.0, 500000.0] regularly spaced with Δy=20833.3
└── Bounded z ∈ [-1000.0, 0.0] regularly spaced with Δz=125.0Model
We built a HydrostaticFreeSurfaceModel with an ImplicitFreeSurface solver. Regarding Coriolis, we use a beta-plane centered at 45° South.
model = HydrostaticFreeSurfaceModel(grid;
coriolis = BetaPlane(latitude = -45),
buoyancy = BuoyancyTracer(),
tracers = :b,
momentum_advection = WENO(),
tracer_advection = WENO())HydrostaticFreeSurfaceModel{CPU, RectilinearGrid}(time = 0 seconds, iteration = 0)
├── grid: 48×48×8 RectilinearGrid{Float64, Periodic, Bounded, Bounded} on CPU with 3×3×3 halo
├── timestepper: QuasiAdamsBashforth2TimeStepper
├── tracers: b
├── closure: Nothing
├── buoyancy: BuoyancyTracer with ĝ = NegativeZDirection()
├── free surface: ImplicitFreeSurface with gravitational acceleration 9.80665 m s⁻²
│ └── solver: FFTImplicitFreeSurfaceSolver
├── advection scheme:
│ ├── momentum: WENO{3, Float64, Float32}(order=5)
│ └── b: WENO{3, Float64, Float32}(order=5)
├── vertical_coordinate: ZCoordinate
└── coriolis: BetaPlane{Float64}We start our simulation from rest with a baroclinically unstable buoyancy distribution. We use ramp(y, Δy), defined below, to specify a front with width Δy and horizontal buoyancy gradient M². We impose the front on top of a vertical buoyancy gradient N² and a bit of noise.
"""
ramp(y, Δy)
Linear ramp from 0 to 1 between -Δy/2 and +Δy/2.
For example:
```
y < -Δy/2 => ramp = 0
-Δy/2 < y < -Δy/2 => ramp = y / Δy
y > Δy/2 => ramp = 1
```
"""
ramp(y, Δy) = min(max(0, y/Δy + 1/2), 1)
N² = 1e-5 # [s⁻²] buoyancy frequency / stratification
M² = 1e-7 # [s⁻²] horizontal buoyancy gradient
Δy = 100kilometers # width of the region of the front
Δb = Δy * M² # buoyancy jump associated with the front
ϵb = 1e-2 * Δb # noise amplitude
bᵢ(x, y, z) = N² * z + Δb * ramp(y, Δy) + ϵb * randn()
set!(model, b=bᵢ)Let's visualize the initial buoyancy distribution.
using CairoMakie
set_theme!(Theme(fontsize = 20))
# Build coordinates with units of kilometers
x, y, z = 1e-3 .* nodes(grid, (Center(), Center(), Center()))
b = model.tracers.b
fig, ax, hm = heatmap(view(b, 1, :, :),
colormap = :deep,
axis = (xlabel = "y [km]",
ylabel = "z [km]",
title = "b(x=0, y, z, t=0)",
titlesize = 24))
Colorbar(fig[1, 2], hm, label = "[m s⁻²]")
figSimulation
Now let's build a Simulation.
simulation = Simulation(model, Δt=20minutes, stop_time=20days)Simulation of HydrostaticFreeSurfaceModel{CPU, RectilinearGrid}(time = 0 seconds, iteration = 0)
├── Next time step: 20 minutes
├── run_wall_time: 0 seconds
├── run_wall_time / iteration: NaN days
├── stop_time: 20 days
├── stop_iteration: Inf
├── wall_time_limit: Inf
├── minimum_relative_step: 0.0
├── callbacks: OrderedDict with 4 entries:
│ ├── stop_time_exceeded => Callback of stop_time_exceeded on IterationInterval(1)
│ ├── stop_iteration_exceeded => Callback of stop_iteration_exceeded on IterationInterval(1)
│ ├── wall_time_limit_exceeded => Callback of wall_time_limit_exceeded on IterationInterval(1)
│ └── nan_checker => Callback of NaNChecker for u on IterationInterval(100)
└── output_writers: OrderedDict with no entriesWe add a TimeStepWizard callback to adapt the simulation's time-step,
conjure_time_step_wizard!(simulation, IterationInterval(20), cfl=0.2, max_Δt=20minutes)Also, we add a callback to print a message about how the simulation is going,
using Printf
wall_clock = Ref(time_ns())
function print_progress(sim)
u, v, w = model.velocities
progress = 100 * (time(sim) / sim.stop_time)
elapsed = (time_ns() - wall_clock[]) / 1e9
@printf("[%05.2f%%] i: %d, t: %s, wall time: %s, max(u): (%6.3e, %6.3e, %6.3e) m/s, next Δt: %s\n",
progress, iteration(sim), prettytime(sim), prettytime(elapsed),
maximum(abs, u), maximum(abs, v), maximum(abs, w), prettytime(sim.Δt))
wall_clock[] = time_ns()
return nothing
end
add_callback!(simulation, print_progress, IterationInterval(100))Diagnostics/Output
Here, we save the buoyancy,
u, v, w = model.velocities
ζ = ∂x(v) - ∂y(u)
B = Average(b, dims=1)
U = Average(u, dims=1)
V = Average(v, dims=1)
filename = "baroclinic_adjustment"
save_fields_interval = 0.5day
slicers = (east = (grid.Nx, :, :),
north = (:, grid.Ny, :),
bottom = (:, :, 1),
top = (:, :, grid.Nz))
for side in keys(slicers)
indices = slicers[side]
simulation.output_writers[side] = JLD2Writer(model, (; b, ζ);
filename = filename * "_$(side)_slice",
schedule = TimeInterval(save_fields_interval),
overwrite_existing = true,
indices)
end
simulation.output_writers[:zonal] = JLD2Writer(model, (; b=B, u=U, v=V);
filename = filename * "_zonal_average",
schedule = TimeInterval(save_fields_interval),
overwrite_existing = true)JLD2Writer scheduled on TimeInterval(12 hours):
├── filepath: baroclinic_adjustment_zonal_average.jld2
├── 3 outputs: (b, u, v)
├── array_type: Array{Float32}
├── including: [:grid, :coriolis, :buoyancy, :closure]
├── file_splitting: NoFileSplitting
└── file size: 0 bytes (file not yet created)Now we're ready to run.
@info "Running the simulation..."
run!(simulation)
@info "Simulation completed in " * prettytime(simulation.run_wall_time)[ Info: Running the simulation...
[ Info: Initializing simulation...
[00.00%] i: 0, t: 0 seconds, wall time: 8.862 seconds, max(u): (0.000e+00, 0.000e+00, 0.000e+00) m/s, next Δt: 20 minutes
[ Info: ... simulation initialization complete (12.167 seconds)
[ Info: Executing initial time step...
[ Info: ... initial time step complete (3.779 seconds).
[06.94%] i: 100, t: 1.389 days, wall time: 14.394 seconds, max(u): (1.209e-01, 1.239e-01, 1.744e-03) m/s, next Δt: 20 minutes
[13.89%] i: 200, t: 2.778 days, wall time: 859.836 ms, max(u): (2.297e-01, 2.011e-01, 2.015e-03) m/s, next Δt: 20 minutes
[20.83%] i: 300, t: 4.167 days, wall time: 904.264 ms, max(u): (2.926e-01, 2.995e-01, 1.868e-03) m/s, next Δt: 20 minutes
[27.78%] i: 400, t: 5.556 days, wall time: 923.161 ms, max(u): (3.910e-01, 4.195e-01, 2.108e-03) m/s, next Δt: 20 minutes
[34.72%] i: 500, t: 6.944 days, wall time: 843.962 ms, max(u): (4.979e-01, 5.832e-01, 2.186e-03) m/s, next Δt: 20 minutes
[41.67%] i: 600, t: 8.333 days, wall time: 884.761 ms, max(u): (7.582e-01, 8.946e-01, 2.961e-03) m/s, next Δt: 20 minutes
[48.61%] i: 700, t: 9.722 days, wall time: 858.798 ms, max(u): (1.245e+00, 1.087e+00, 3.927e-03) m/s, next Δt: 20 minutes
[55.56%] i: 800, t: 11.111 days, wall time: 1.121 seconds, max(u): (1.343e+00, 1.120e+00, 5.626e-03) m/s, next Δt: 20 minutes
[62.50%] i: 900, t: 12.500 days, wall time: 2.144 seconds, max(u): (1.320e+00, 1.132e+00, 5.085e-03) m/s, next Δt: 20 minutes
[69.44%] i: 1000, t: 13.889 days, wall time: 1.407 seconds, max(u): (1.490e+00, 1.040e+00, 4.472e-03) m/s, next Δt: 20 minutes
[76.39%] i: 1100, t: 15.278 days, wall time: 1.296 seconds, max(u): (1.365e+00, 9.538e-01, 2.625e-03) m/s, next Δt: 20 minutes
[83.33%] i: 1200, t: 16.667 days, wall time: 1.047 seconds, max(u): (1.335e+00, 8.977e-01, 3.105e-03) m/s, next Δt: 20 minutes
[90.28%] i: 1300, t: 18.056 days, wall time: 909.227 ms, max(u): (1.416e+00, 1.100e+00, 2.438e-03) m/s, next Δt: 20 minutes
[97.22%] i: 1400, t: 19.444 days, wall time: 849.118 ms, max(u): (1.250e+00, 1.122e+00, 2.573e-03) m/s, next Δt: 20 minutes
[ Info: Simulation is stopping after running for 31.234 seconds.
[ Info: Simulation time 20 days equals or exceeds stop time 20 days.
[ Info: Simulation completed in 31.277 secondsVisualization
All that's left is to make a pretty movie. Actually, we make two visualizations here. First, we illustrate how to make a 3D visualization with Makie's Axis3 and Makie.surface. Then we make a movie in 2D. We use CairoMakie in this example, but note that using GLMakie is more convenient on a system with OpenGL, as figures will be displayed on the screen.
using CairoMakieThree-dimensional visualization
We load the saved buoyancy output on the top, north, and east surface as FieldTimeSerieses.
filename = "baroclinic_adjustment"
sides = keys(slicers)
slice_filenames = NamedTuple(side => filename * "_$(side)_slice.jld2" for side in sides)
b_timeserieses = (east = FieldTimeSeries(slice_filenames.east, "b"),
north = FieldTimeSeries(slice_filenames.north, "b"),
top = FieldTimeSeries(slice_filenames.top, "b"))
B_timeseries = FieldTimeSeries(filename * "_zonal_average.jld2", "b")
times = B_timeseries.times
grid = B_timeseries.grid48×48×8 RectilinearGrid{Float64, Periodic, Bounded, Bounded} on CPU with 3×3×3 halo
├── Periodic x ∈ [0.0, 1.0e6) regularly spaced with Δx=20833.3
├── Bounded y ∈ [-500000.0, 500000.0] regularly spaced with Δy=20833.3
└── Bounded z ∈ [-1000.0, 0.0] regularly spaced with Δz=125.0We build the coordinates. We rescale horizontal coordinates to kilometers.
xb, yb, zb = nodes(b_timeserieses.east)
xb = xb ./ 1e3 # convert m -> km
yb = yb ./ 1e3 # convert m -> km
Nx, Ny, Nz = size(grid)
x_xz = repeat(x, 1, Nz)
y_xz_north = y[end] * ones(Nx, Nz)
z_xz = repeat(reshape(z, 1, Nz), Nx, 1)
x_yz_east = x[end] * ones(Ny, Nz)
y_yz = repeat(y, 1, Nz)
z_yz = repeat(reshape(z, 1, Nz), grid.Ny, 1)
x_xy = x
y_xy = y
z_xy_top = z[end] * ones(grid.Nx, grid.Ny)Then we create a 3D axis. We use zonal_slice_displacement to control where the plot of the instantaneous zonal average flow is located.
fig = Figure(size = (1600, 800))
zonal_slice_displacement = 1.2
ax = Axis3(fig[2, 1],
aspect=(1, 1, 1/5),
xlabel = "x (km)",
ylabel = "y (km)",
zlabel = "z (m)",
xlabeloffset = 100,
ylabeloffset = 100,
zlabeloffset = 100,
limits = ((x[1], zonal_slice_displacement * x[end]), (y[1], y[end]), (z[1], z[end])),
elevation = 0.45,
azimuth = 6.8,
xspinesvisible = false,
zgridvisible = false,
protrusions = 40,
perspectiveness = 0.7)Axis3 with 0 plots:We use data from the final savepoint for the 3D plot. Note that this plot can easily be animated by using Makie's Observable. To dive into Observables, check out Makie.jl's Documentation.
n = length(times)41Now let's make a 3D plot of the buoyancy and in front of it we'll use the zonally-averaged output to plot the instantaneous zonal-average of the buoyancy.
b_slices = (east = interior(b_timeserieses.east[n], 1, :, :),
north = interior(b_timeserieses.north[n], :, 1, :),
top = interior(b_timeserieses.top[n], :, :, 1))
# Zonally-averaged buoyancy
B = interior(B_timeseries[n], 1, :, :)
clims = 1.1 .* extrema(b_timeserieses.top[n][:])
kwargs = (colorrange=clims, colormap=:deep, shading=NoShading)
surface!(ax, x_yz_east, y_yz, z_yz; color = b_slices.east, kwargs...)
surface!(ax, x_xz, y_xz_north, z_xz; color = b_slices.north, kwargs...)
surface!(ax, x_xy, y_xy, z_xy_top; color = b_slices.top, kwargs...)
sf = surface!(ax, zonal_slice_displacement .* x_yz_east, y_yz, z_yz; color = B, kwargs...)
contour!(ax, y, z, B; transformation = (:yz, zonal_slice_displacement * x[end]),
levels = 15, linewidth = 2, color = :black)
Colorbar(fig[2, 2], sf, label = "m s⁻²", height = Relative(0.4), tellheight=false)
title = "Buoyancy at t = " * string(round(times[n] / day, digits=1)) * " days"
fig[1, 1:2] = Label(fig, title; fontsize = 24, tellwidth = false, padding = (0, 0, -120, 0))
rowgap!(fig.layout, 1, Relative(-0.2))
colgap!(fig.layout, 1, Relative(-0.1))
save("baroclinic_adjustment_3d.png", fig)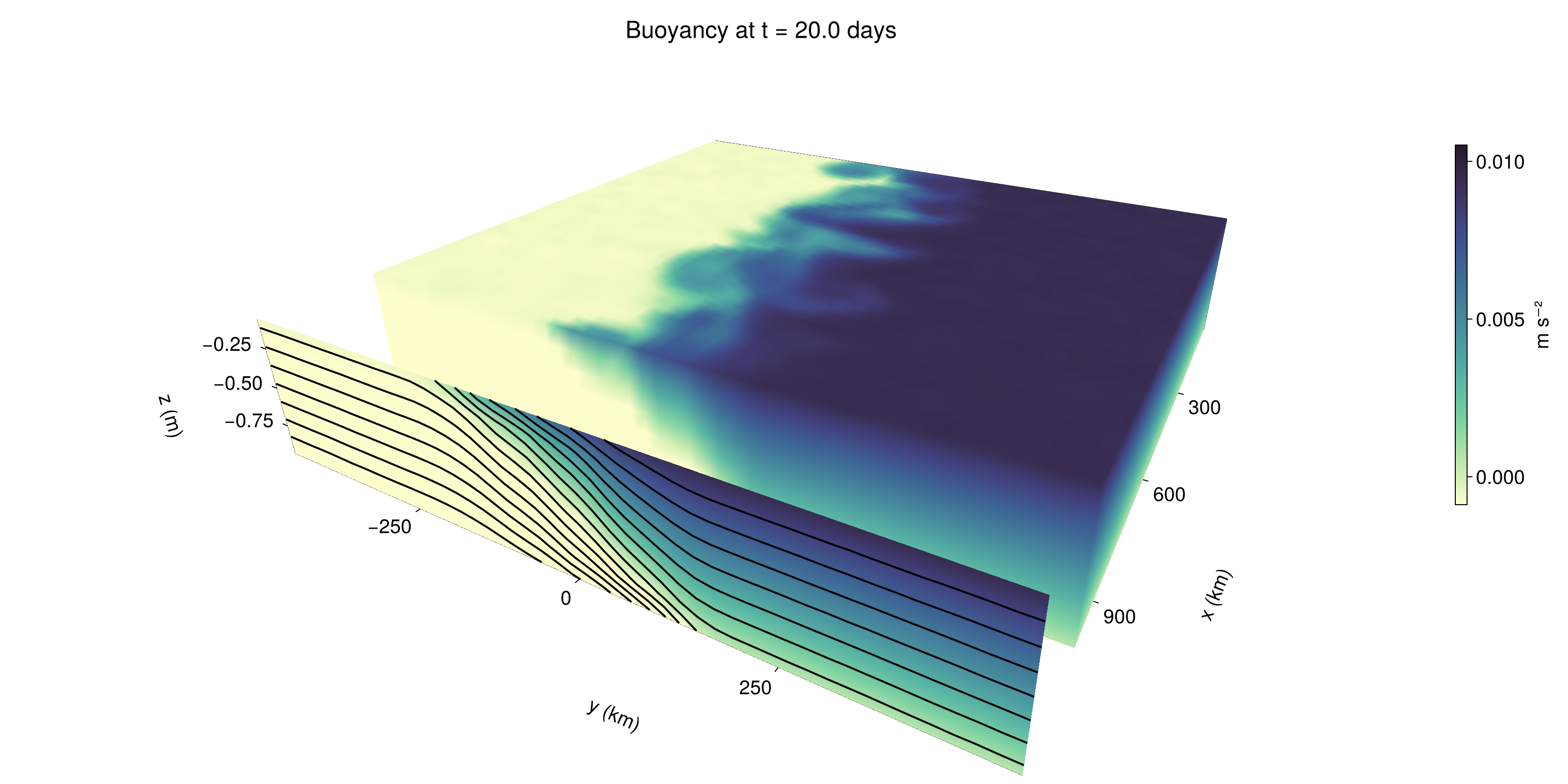
Two-dimensional movie
We make a 2D movie that shows buoyancy FieldTimeSeries and extract the additional coordinates we'll need for plotting
ζ_timeseries = FieldTimeSeries(slice_filenames.top, "ζ")
U_timeseries = FieldTimeSeries(filename * "_zonal_average.jld2", "u")
B_timeseries = FieldTimeSeries(filename * "_zonal_average.jld2", "b")
V_timeseries = FieldTimeSeries(filename * "_zonal_average.jld2", "v")
xζ, yζ, zζ = nodes(ζ_timeseries)
yv = ynodes(V_timeseries)
xζ = xζ ./ 1e3 # convert m -> km
yζ = yζ ./ 1e3 # convert m -> km
yv = yv ./ 1e3 # convert m -> km-500.0:20.833333333333332:500.0Next, we set up a plot with 4 panels. The top panels are large and square, while the bottom panels get a reduced aspect ratio through rowsize!.
fig = Figure(size=(1800, 1000))
axb = Axis(fig[1, 2], xlabel="x (km)", ylabel="y (km)", aspect=1)
axζ = Axis(fig[1, 3], xlabel="x (km)", ylabel="y (km)", aspect=1, yaxisposition=:right)
axu = Axis(fig[2, 2], xlabel="y (km)", ylabel="z (m)")
axv = Axis(fig[2, 3], xlabel="y (km)", ylabel="z (m)", yaxisposition=:right)
rowsize!(fig.layout, 2, Relative(0.3))To prepare a plot for animation, we index the timeseries with an Observable,
n = Observable(1)
b_top = @lift interior(b_timeserieses.top[$n], :, :, 1)
ζ_top = @lift interior(ζ_timeseries[$n], :, :, 1)
U = @lift interior(U_timeseries[$n], 1, :, :)
V = @lift interior(V_timeseries[$n], 1, :, :)
B = @lift interior(B_timeseries[$n], 1, :, :)Observable([-0.009359133429825306 -0.00812368094921112 -0.006887644063681364 -0.005612972192466259 -0.004392298404127359 -0.003117109416052699 -0.001858846633695066 -0.0006257317727431655; -0.009385657496750355 -0.008114910684525967 -0.0068826437927782536 -0.005624826066195965 -0.004372659604996443 -0.0031191986054182053 -0.0018679380882531404 -0.0006154820439405739; -0.009382609277963638 -0.008118750527501106 -0.006875529885292053 -0.00562229473143816 -0.004389068577438593 -0.0031363130547106266 -0.0018823074642568827 -0.0006291751633398235; -0.009396238252520561 -0.008137643337249756 -0.006872963160276413 -0.0056344508193433285 -0.004368090070784092 -0.003124195383861661 -0.0018658916233107448 -0.0006509340018965304; -0.009373386390507221 -0.008141309954226017 -0.006898436229676008 -0.005623212084174156 -0.004369232803583145 -0.0031412148382514715 -0.0018752976320683956 -0.0006229709833860397; -0.009342999197542667 -0.008127513341605663 -0.006871216464787722 -0.005623199045658112 -0.004381462000310421 -0.003111507510766387 -0.0018774009076878428 -0.0006240666261874139; -0.00940277986228466 -0.008088666014373302 -0.00687830476090312 -0.005642759148031473 -0.0043691061437129974 -0.0031357731204479933 -0.0018779977690428495 -0.0006114757852628827; -0.009363423101603985 -0.008132143877446651 -0.006862174719572067 -0.00559989083558321 -0.004372556693851948 -0.00315172434784472 -0.0018802075646817684 -0.0006323513225652277; -0.009374127723276615 -0.00811651349067688 -0.006855000276118517 -0.005621314514428377 -0.004341508727520704 -0.0031287551391869783 -0.0018760706298053265 -0.0006223986856639385; -0.009362866170704365 -0.008106926456093788 -0.006886397022753954 -0.005622498691082001 -0.004371571820229292 -0.0031188339926302433 -0.0018694315804168582 -0.0006329355528578162; -0.009385637007653713 -0.008142033591866493 -0.00686337286606431 -0.005610644351691008 -0.00436315918341279 -0.003118698950856924 -0.0018572928383946419 -0.0006206324906088412; -0.009378260932862759 -0.008139369077980518 -0.006843898445367813 -0.005625503603368998 -0.004338220693171024 -0.0031021093018352985 -0.0018720212392508984 -0.0006333422497846186; -0.00936328899115324 -0.008125337772071362 -0.006884688511490822 -0.005612085107713938 -0.00434535089880228 -0.0031217136420309544 -0.001883418415673077 -0.0006179129704833031; -0.009373066946864128 -0.008143617771565914 -0.006870725657790899 -0.005624591372907162 -0.0043680802918970585 -0.0031235588248819113 -0.0018938201246783137 -0.000630145485047251; -0.009370485320687294 -0.008133219555020332 -0.006868159398436546 -0.005648565012961626 -0.004375993739813566 -0.003130938857793808 -0.0018869206542149186 -0.0006223592208698392; -0.009366430342197418 -0.008153781294822693 -0.006881895940750837 -0.005645676515996456 -0.004376196302473545 -0.00313364015892148 -0.0018457944970577955 -0.0006074339034967124; -0.009362840093672276 -0.008136124350130558 -0.006890094839036465 -0.005613446235656738 -0.0043736714869737625 -0.0031218938529491425 -0.001875598100014031 -0.0005753047880716622; -0.009377103298902512 -0.008137114346027374 -0.006869750563055277 -0.005613009445369244 -0.004368535242974758 -0.0031267907470464706 -0.0018716342747211456 -0.0006155339651741087; -0.009365211240947247 -0.008119330741465092 -0.0068611628375947475 -0.00563859473913908 -0.004398012533783913 -0.003137597581371665 -0.0018636897439137101 -0.0006232837913557887; -0.009373864158987999 -0.008139301091432571 -0.006871933117508888 -0.0056021795608103275 -0.004377895034849644 -0.003134669503197074 -0.001874146400950849 -0.0006027621566317976; -0.009387359023094177 -0.008122810162603855 -0.00687003880739212 -0.0055997841991484165 -0.004390757530927658 -0.0031497240997850895 -0.0018667674157768488 -0.0006315585342235863; -0.009400763548910618 -0.008123325183987617 -0.006867910269647837 -0.0056228418834507465 -0.0043689352460205555 -0.0031025423668324947 -0.0019004751229658723 -0.0006436920375563204; -0.007497962098568678 -0.006267086137086153 -0.004969359841197729 -0.003751518903300166 -0.0025059340987354517 -0.0012753552291542292 1.5468323908862658e-5 0.001251854351721704; -0.00541406124830246 -0.004159293137490749 -0.002910604700446129 -0.0016256177332252264 -0.00041484463145025074 0.0008445536950603127 0.0020659975707530975 0.003338235430419445; -0.0032901582308113575 -0.0020800044294446707 -0.0008393650641664863 0.000400648161303252 0.0016521764919161797 0.0029234245885163546 0.004165601450949907 0.005418123211711645; -0.0012556061847135425 -1.8122280380339362e-5 0.001233158865943551 0.002519880188629031 0.003732440760359168 0.004993794951587915 0.006238317582756281 0.007519937586039305; 0.0006277432548813522 0.001877636881545186 0.0031450565438717604 0.004365346860140562 0.005622479598969221 0.006876819301396608 0.008138147182762623 0.009400474838912487; 0.0006301752873696387 0.001886544399894774 0.0031544521916657686 0.004388913046568632 0.005629288498312235 0.006879190448671579 0.008135109208524227 0.009398916736245155; 0.000628949492238462 0.0018767726141959429 0.003126031020656228 0.00435793399810791 0.005627578590065241 0.0068986075930297375 0.008143726736307144 0.00939362496137619; 0.0006161114433780313 0.001872982014901936 0.003129094373434782 0.004392250906676054 0.005628467537462711 0.006880093831568956 0.008124131709337234 0.00935440044850111; 0.0006390219205059111 0.001871234504505992 0.0031255409121513367 0.004352213349193335 0.005611718166619539 0.00686672143638134 0.008100675418972969 0.009396331384778023; 0.0006092846742831171 0.0018686344847083092 0.003108177799731493 0.004371221177279949 0.005643206182867289 0.006864042021334171 0.008129113353788853 0.009357575327157974; 0.0006142235361039639 0.0018923879833891988 0.003108890261501074 0.0043784440495073795 0.005596166010946035 0.006861128844320774 0.008128813467919827 0.009377259761095047; 0.0006370479823090136 0.0018991308752447367 0.0031311626080423594 0.004388940520584583 0.005645280238240957 0.006869713310152292 0.008128794841468334 0.009377975948154926; 0.0006094860145822167 0.0018839157419279218 0.0031498740427196026 0.004380751401185989 0.0056142681278288364 0.006871548481285572 0.008125394582748413 0.009379691444337368; 0.0006325605791062117 0.0018869113409891725 0.0031259111128747463 0.004353533964604139 0.005627073347568512 0.006880634929984808 0.008129487745463848 0.009364933706820011; 0.0006397597026079893 0.0018854821100831032 0.0031272126361727715 0.004384187515825033 0.005618694704025984 0.00685417465865612 0.008138210512697697 0.009376650676131248; 0.000621538725681603 0.0018628968391567469 0.0031266645528376102 0.00436983210965991 0.005643346346914768 0.006868122145533562 0.008125653490424156 0.009357270784676075; 0.0006199812050908804 0.001897216192446649 0.0031238202936947346 0.004367181099951267 0.005613934714347124 0.006871039047837257 0.008134829811751842 0.009376189671456814; 0.000632559007499367 0.0018742268439382315 0.0031128579284995794 0.004385008011013269 0.0056205978617072105 0.0068815043196082115 0.008115083910524845 0.009366856887936592; 0.0006009367643855512 0.0018557654693722725 0.003111886326223612 0.004374169744551182 0.005596277303993702 0.0068721892312169075 0.00813806802034378 0.009385846555233002; 0.0006295129423961043 0.0018708071438595653 0.0031394411344081163 0.004375811200588942 0.005617818329483271 0.006875690072774887 0.008128034882247448 0.009395286440849304; 0.0006375876837410033 0.0018771733157336712 0.0031247821170836687 0.004376022145152092 0.005611030384898186 0.006887687835842371 0.008152051828801632 0.009359029121696949; 0.0006111768307164311 0.0018929592333734035 0.0031412423122674227 0.004373006988316774 0.005606588441878557 0.00687739672139287 0.008105553686618805 0.009389991872012615; 0.0006672492600046098 0.0018849197076633573 0.0031543096993118525 0.004369175992906094 0.005636714864522219 0.006862018723040819 0.008122554048895836 0.009373343549668789; 0.0006148535758256912 0.0018619688926264644 0.0031109570991247892 0.0043782261200249195 0.005639717448502779 0.006868751719594002 0.00811187643557787 0.009369689971208572; 0.000627342436928302 0.0019133654423058033 0.0031232452020049095 0.004406054504215717 0.005643337499350309 0.006865829229354858 0.008154616691172123 0.009379023686051369; 0.0006353645585477352 0.0018539638258516788 0.0031380741856992245 0.004372351802885532 0.00561935268342495 0.006865436676889658 0.00812129583209753 0.009358607232570648])and then build our plot:
hm = heatmap!(axb, xb, yb, b_top, colorrange=(0, Δb), colormap=:thermal)
Colorbar(fig[1, 1], hm, flipaxis=false, label="Surface b(x, y) (m s⁻²)")
hm = heatmap!(axζ, xζ, yζ, ζ_top, colorrange=(-5e-5, 5e-5), colormap=:balance)
Colorbar(fig[1, 4], hm, label="Surface ζ(x, y) (s⁻¹)")
hm = heatmap!(axu, yb, zb, U; colorrange=(-5e-1, 5e-1), colormap=:balance)
Colorbar(fig[2, 1], hm, flipaxis=false, label="Zonally-averaged U(y, z) (m s⁻¹)")
contour!(axu, yb, zb, B; levels=15, color=:black)
hm = heatmap!(axv, yv, zb, V; colorrange=(-1e-1, 1e-1), colormap=:balance)
Colorbar(fig[2, 4], hm, label="Zonally-averaged V(y, z) (m s⁻¹)")
contour!(axv, yb, zb, B; levels=15, color=:black)Finally, we're ready to record the movie.
frames = 1:length(times)
record(fig, filename * ".mp4", frames, framerate=8) do i
n[] = i
endJulia version and environment information
This example was executed with the following version of Julia:
using InteractiveUtils: versioninfo
versioninfo()Julia Version 1.12.4
Commit 01a2eadb047 (2026-01-06 16:56 UTC)
Build Info:
Official https://julialang.org release
Platform Info:
OS: Linux (x86_64-linux-gnu)
CPU: 128 × AMD EPYC 9374F 32-Core Processor
WORD_SIZE: 64
LLVM: libLLVM-18.1.7 (ORCJIT, znver4)
GC: Built with stock GC
Threads: 1 default, 1 interactive, 1 GC (on 128 virtual cores)
Environment:
LD_LIBRARY_PATH =
JULIA_PKG_SERVER_REGISTRY_PREFERENCE = eager
JULIA_DEPOT_PATH = /var/lib/buildkite-agent/.julia-oceananigans
JULIA_PROJECT = /var/lib/buildkite-agent/Oceananigans.jl-29835/docs/
JULIA_VERSION = 1.12.4
JULIA_LOAD_PATH = @:@v#.#:@stdlib
JULIA_VERSION_ENZYME = 1.10.10
JULIA_PYTHONCALL_EXE = /var/lib/buildkite-agent/Oceananigans.jl-29835/docs/.CondaPkg/.pixi/envs/default/bin/python
JULIA_DEBUG = LiterateThese were the top-level packages installed in the environment:
import Pkg
Pkg.status()Status `~/Oceananigans.jl-29835/docs/Project.toml`
[79e6a3ab] Adapt v4.5.0
[052768ef] CUDA v5.9.7
[13f3f980] CairoMakie v0.15.9
[e30172f5] Documenter v1.17.0
[daee34ce] DocumenterCitations v1.4.1
[4710194d] DocumenterVitepress v0.3.2
[033835bb] JLD2 v0.6.3
[63c18a36] KernelAbstractions v0.9.40
[98b081ad] Literate v2.21.0
[da04e1cc] MPI v0.20.23
[85f8d34a] NCDatasets v0.14.12
[9e8cae18] Oceananigans v0.105.3 `..`
[f27b6e38] Polynomials v4.1.1
[6038ab10] Rotations v1.7.1
[d496a93d] SeawaterPolynomials v0.3.10
[09ab397b] StructArrays v0.7.2
[bdfc003b] TimesDates v0.3.3
[2e0b0046] XESMF v0.1.6
[b77e0a4c] InteractiveUtils v1.11.0
[37e2e46d] LinearAlgebra v1.12.0
[44cfe95a] Pkg v1.12.1This page was generated using Literate.jl.